Broad genome analysis shows yeasts evolving by subtraction
An unprecedented comparison of hundreds of species of yeasts has helped geneticists brew up an expansive picture of their evolution over the last hundreds of millions of years, including an analysis of the way they evolved individual appetites for particular food sources that may be a boon to biofuels research.
A team of researchers led by labs at the University of Wisconsin–Madison and Vanderbilt University sequenced and compared the genomes of 332 species of budding yeasts, all members of a subphylum of yeasts that multiply by producing daughter cells from buds on their surface. More than 200 of the yeast types had their genomes catalogued for the first time for the study, which was published today (Nov. 8, 2018) by the journal Cell.

Blastobotrys americana, seen here growing in colonies in a dish in a UW–Madison lab, was among 332 yeast species to have its genome sequenced and analyzed by researchers in a study that described their evolution and specialization. Courtesy of Dana Opulente
Even in an era of big-data comparisons of DNA — when studies often involve analyzing the genes of many people or many fruit flies at a time — the yeast study is a step into new territory.
“This is the first large genome project like this that actually looks at hundreds of different eukaryotic species, not different individuals or isolates of the same species,” says Chris Todd Hittinger, a UW–Madison genetics professor and one of the senior authors of the study. “Budding yeasts, despite their phenotypic similarity, are very different from one another genetically. They’re as different from one another as all animals or all plants are from one another.”
Collecting such a deep pool of yeast types gave researchers enough information to use comparisons of the shifting genetics to redraw the budding yeast family tree into a dozen major branches and paint a detailed picture of their past.
“By having a very broad swath of biodiversity, it allows us to reconstruct the evolutionary processes through time,” Hittinger says. “That’s what allows us to make the inference that much of budding yeast evolution occurred through the process of reductive evolution, where you have a relatively metabolically complex common ancestor losing traits through time.”
That common ancestor is an ancient yeast whose progeny began about 400 million years ago to differentiate into the study’s hundreds of yeast species. That includes a fork about 5 million years ago that gave the world Saccharomyces cerevisiae, the yeast that brewers, bakers and wine-makers rely on to put alcohol in their bottles and rise in their bread.
The original Saccharomycotina was probably more complex than its descendants in an important regard.
The researchers examined their yeasts for 45 traits representing their ability to process a variety of yeast foods — different sources of carbon and nitrogen necessary to store energy and build cells. Tracking back the evolutionary paths of modern yeasts suggests the common ancestor yeast had a metabolism that could work with a varied diet.
“We have a more consistent picture now of the variations of carbon and nitrogen sources across the modern species,” says Dana Opulente, a postdoctoral researcher in Hittinger’s lab who redid much of the trait-testing work of a century of yeast researchers for the Cell study. “They show us that this ancestor yeast would have been able to use a wider array of sugars than modern budding yeasts.”
Modern yeasts have narrowed their appetites in a process called reductive evolution, losing quite a few of those 45 traits as they specialized to flourish in their particular niches.
“To pick on the model budding yeast, S. cerevisiae has one of the more reduced genomes,” Hittinger says. “It lacks many of the metabolic capabilities that other budding yeasts have.”
Describing yeast metabolism and the related genes is important for bioenergy production, which relies on yeast and other microbes to turn plant cells into useful, renewable fuels or compounds that can be further refined into fuels.
“Plant biomass is very complex and breaks down into many different kinds of sugars and other molecules,” says Hittinger, whose lab is affiliated with the UW–Madison-based, Department of Energy-funded Great Lakes Bioenergy Research Center. “Ultimately what you want is a microbe that’s going to be able to eat as many of them as possible and turn them into something useful.”
Modern yeasts may have become pickier eaters, but the new analysis of their genomes can help biofuels researchers pinpoint helpful skills among a daunting number of candidate yeasts.
“If they want a yeast that’s good at fermenting a sugar like xylose, they don’t have to think, ‘Oh, no, there’s more than 1,000 kinds of yeasts. Where do I start?’” says Jacek Kominek, a Hittinger lab geneticist and (with Opulente) one of the lead authors of the Cell study. “This project gives them a foundation, another way to decide where the most interesting ones are.”
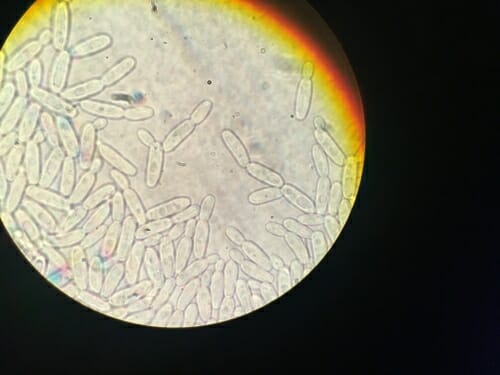
An analysis of the genomes of 332 yeast species — like Pichia exigua, seen here through a microscope — may help biofuels researchers identify helpful microbes for turning plant material into new, renewable energy sources. Courtesy of Dana Opulente
Better still, the new study’s data can help scientists delve further into the workings of those interesting yeasts to pick apart the genetic differences between a yeast that happily munches on a particular plant molecule and one that struggles with the same compound.
“Maybe they find an extra gene that makes one yeast more useful, and maybe they can put that gene into a yeast they’re used to using already, like S. cerevisiae,” Opulente says.
And maybe they can pair advantageous inputs with rewarding outputs.
“One of the most exciting things about the genetic diversity we’re tapping into here is seeing that many of the yeasts that actually have the broadest appetite for different carbon sources also produce some of the most interesting energy-storage molecules,” Hittinger says.
Some of them can pack away more than half their weight as oily lipids that are potential biodiesel precursors.
“Because these species have such a broad appetite and because they put their carbon in other places than in ethanol, they’re pushing carbon down different pathways,” Hittinger says. “In some cases, it’s right toward different fuels, fuel precursors or higher-value chemicals.”
Collaborators on the study include researchers from the U.S. Department of Agriculture, South China Agricultural University, New York University, Purdue University, RIKEN BioResource Research Center and Center for Life Science Technologies in Japan, University of Ljubljana in Slovenia, Universidad Nacional del Comahue in Argentina, Universidade Federal de Minas Gerais in Brazil, and Westerdijk Fungal Biodiversity Institute.
Support for this study came from the National Science Foundation (grants DEB-1442113, DEB-1442148, IOS-1401682, and DGE-1256259), the Department of Energy (DE-FC02-07ER64494 and DE-SC0018409) and the National Institutes of Health (5T32GM007133).





