Research on viral junk, quicker drug testing could help outflank coronaviruses
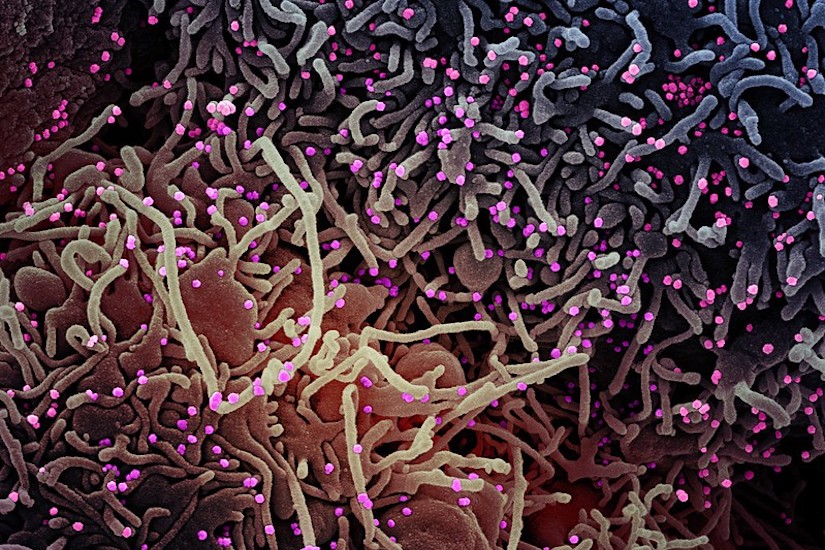
Illustration of a cell being infected by COVID-19 particles (shown in pink). Researchers hope to better understand how coronaviruses enter cells, spread, and cause varying immune responses in different individuals. NIAID/NIH
Viruses like novel coronavirus can be sloppy multipliers, leaving lots of junk particles around infected cells during reproduction. That junk may help humans fight illness.
Most models of diseases like COVID-19 are based on purified virus stock. But in nature, many viruses are not particularly pure. When RNA viruses, which include coronaviruses, replicate, the process produces lots of partial viruses, mutations and other “junk.” These are also known as defective interfering particles, and can no longer replicate and spread themselves.
On a potentially positive note, these particles can tie up resources, making it more difficult for an active virus to spread from cell to cell in its host.
John Yin, a University of Wisconsin–Madison chemical engineering professor — along with colleagues from chemical and biological engineering and the interdisciplinary Wisconsin Institute for Discovery, where Yin is a faculty member — plans to find out if these defective interfering particles, documented previously in mouse coronaviruses, exist in human coronaviruses as well. If that’s the case, their next step may be to examine how viruses spread as a mixture of active and defective virus strains and how those particles interact with one another and with their host cells.
Yin has secured support from the National Science Foundation through a Rapid Response Research (RAPID) grant and an Early-Concept Grant for Exploratory Research (EAGER). In March, Congress appropriated $75 million to NSF for RAPID and EAGER grants to fund projects that prevent, prepare for and respond to coronavirus, domestically or internationally.
For the RAPID project, the team will take an ecological view of coronaviruses to better understand how they enter cells, spread and cause varying immune responses in different individuals.
By understanding the relationships between virus, defective interfering particles and hosts, it may be possible to manipulate the environment to control or slow the spread of the virus. The idea, Yin says, is to look at the viral ecosystem in ways similar to the microbiome, or the variety of bacteria in the human digestive tract. Researchers are beginning to understand that the diversity and ratio of that bacteria can have big impacts on disease and health.
“That same kind of ecological thinking is much more slowly entering virology,” says Yin. “One aspect of it is an appreciation for the diversity of the products that come out of an infected cell. It’s not just a viable virus that wants to spread the infection, but there are also other products like defective particles that might actually limit the infection.”
To understand this complex system, the team will use what is known about the virus’s growth and spread to build mathematical models incorporating the ways the virus enters cells and expresses genes. The team will conduct experiments on human coronaviruses, but not the novel coronavirus causing the COVID-19 pandemic.
The EAGER grant will support the development of a new kind of testing for antiviral drugs.
In the current gold-standard test, a plaque assay, researchers release a virus onto a dish of cells overlaid with a layer of semi-solid agar to force the virus to spread only to neighboring cells. But Yin, along with former graduate student Ying Zhu, found that by replacing the agar with a thin layer of flowing liquid medium, they could reduce testing time from three to five days to just a day or two while increasing the tests’ sensitivity.
Yin will collaborate with chemical and biological engineering Professor Michael Graham to determine the best conditions for testing drugs that may be effective against COVID-19, bringing more of the chemical engineer’s unique perspective of to virology and the fight against COVID-19.

