New tool for assessing heart muscle cells helps unlock their potential
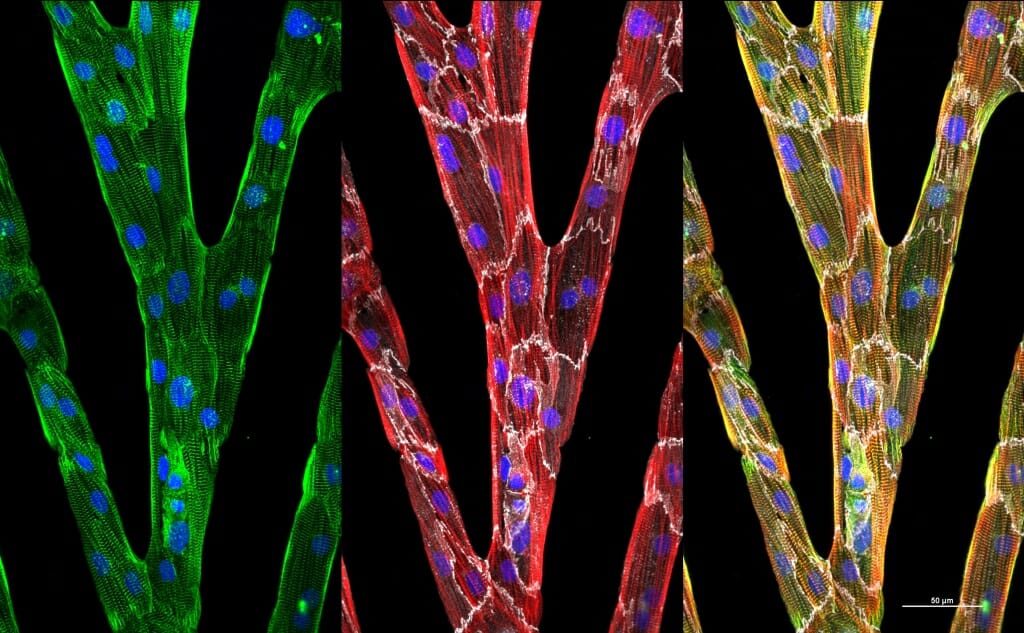
A new image analysis technique designed by UW–Madison engineers can quantify the organization and alignment of heart muscle cells (like these cell groups, in which individual cells are marked by fine white outlines) and help them grow more useful muscle tissue from stem cells. Image courtesy of Brett N. Napiwocki
Heart muscle cells made from stem cells could be used to screen drugs or develop cell-based therapies for heart disease. But these cells, called cardiomyocytes, are often immature, disorganized and unable to behave together like working muscle tissue.
University of Wisconsin–Madison researchers have created a new way to study how fully capable heart cells organize themselves into muscle with rod-like units called myofibrils which can contract together to pump blood. What they learn could help them steer cardiomyocytes into more useful stages of development.
“We want to produce mature cardiomyocytes with internal structures that are nicely organized and aligned, which allows them to work in a coordinated way with their neighboring cells to generate functional muscle behavior,” says Wendy Crone, a University of Wisconsin–Madison professor of engineering physics.
A team led by Crone has created a powerful tool to help assess what experimental factors achieve this goal. The researchers developed an algorithm for image analysis that quantifies the organization and alignment of structures within the myofibrils of cardiomyocytes with considerable accuracy. The team described its advance in a paper published in May in the Journal of Applied Physics.
Crone says this new technique, called the scanning gradient Fourier transform (SGFT) method, gives researchers a valuable tool for evaluating cells and determining the most effective interventions for coaxing immature cardiomyocytes to become mature cells with highly organized internal structures.
The UW–Madison researchers’ method has a key advantage over other image analysis techniques: It is adept at analyzing cells that have some level of organization to their internal structures but fall short of being extremely well-organized.
“We started this project because we needed a way to quantify the organization of cells in these intermediate states,” Crone says. “It’s particularly important for our research because we want to be able to look at cultured cells as they mature—where they start out being very disorganized and then end up developing a much more organized state over time.”
Crone’s research focuses on culturing cardiomyocytes and other cell types derived from stem cells, so writing code for image analysis was a unique undertaking for her research group. She credits former graduate student Max Salick, the first author on the paper, for developing a deep understanding of image analysis and writing the primary code for the algorithm.
The usefulness of the new algorithm also extends beyond cardiomyocytes. Researchers can use it with a variety of imaging techniques and different cell types to effectively assess organization in general, Crone says.
For example, she already has collaborated with colleague Randolph Ashton, a UW–Madison associate professor of biomedical engineering, to demonstrate that the algorithm can quantify the organization of neural rosettes, which are early stage neurons derived from stem cells.
In the new study, the researchers also reported using their method to analyze a breast cancer tissue sample. They showed their technique was able to clearly detect the organization of the collagen fibers within the tissue.
“That’s important because if the collagen in breast tissue is very well organized, it’s a strong indicator of the malignancy of the tumor,” Crone says. “So organization can be good or bad depending on the situation, and our method allows us to quantify it in a variety of different scenarios.
This research was supported in part by grants from the National Institutes of Health.
Enjoy this story?
Read more news from the College of EngineeringTags: College of Engineering, health, research, stem cells
