Developing drug resistance may be a matter of diversity for tuberculosis
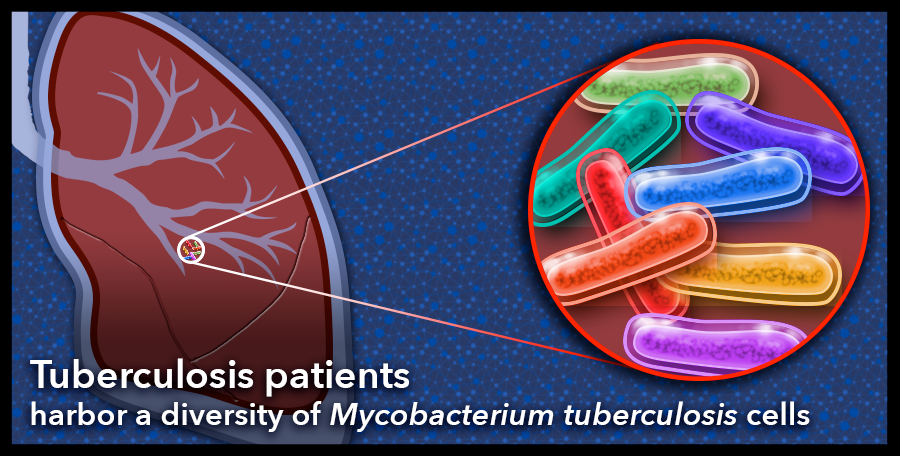
The bacterial cells that infect a person with tuberculosis are highly diverse in individual patients with severe disease. The human body plays host to Mycobacterium tuberculosis cells that adapt readily to their environments and vary genetically. This diversity is especially prevalent in the genes that code for the bacterial cell envelope and may be important for host-pathogen interactions, including the development of drug resistance. Graphic: Sue Medaris
To a microbe, the human body is a vast environment, full of resources and opportunities, dangers and threats. In the world of bacteria, it’s thrive or fail to survive. Evolve or go extinct.
Caitlin Pepperell, professor of medicine and medical microbiology and immunology at the University of Wisconsin–Madison, wants to know more about microbes and their interactions with their human environments, particularly as more pathogens evolve resistance to the drugs we use to combat them.
In a new study recently published in PLoS Pathogens, Pepperell and colleagues at UW–Madison probed the bacteria that causes tuberculosis, Mycobacterium tuberculosis, to learn more about how individual bacterial cells change and adapt while in the human body.
“If you understand what evolutionary tricks the bug has, you are in a better position to tackle the emergence of resistance,” says Pepperell, who is also an infectious disease physician. “What you want to do when you treat an infection is cause extinction, and the more you know about how the bacterial population evolves, the smarter you can be about making it go extinct.”
The researchers wanted to know: How diverse are the bacterial cells that reside within a single person? Where in the bacterial genetic code are the changes that lead to adaptation most extreme?
“Really, we set out to find patterns of diversity and what we could learn from them,” Pepperell says.
To do this, they needed data in the form of the long strings of letters that make up the genetic code of individual tuberculosis bacterial cells harbored by patients with the disease. Tuberculosis bacteria primarily infect the lungs but can be found anywhere in the body, including the brain and kidneys.
The researchers found data collected from five patients in three previous studies, all of whom had severe disease and developed resistance to at least one common tuberculosis drug. The available data had been collected over time, as the patients underwent treatment and their disease progressed. Two ultimately died of tuberculosis, which was once the leading cause of death in the United States and is still often fatal, especially among people who develop drug resistant disease.
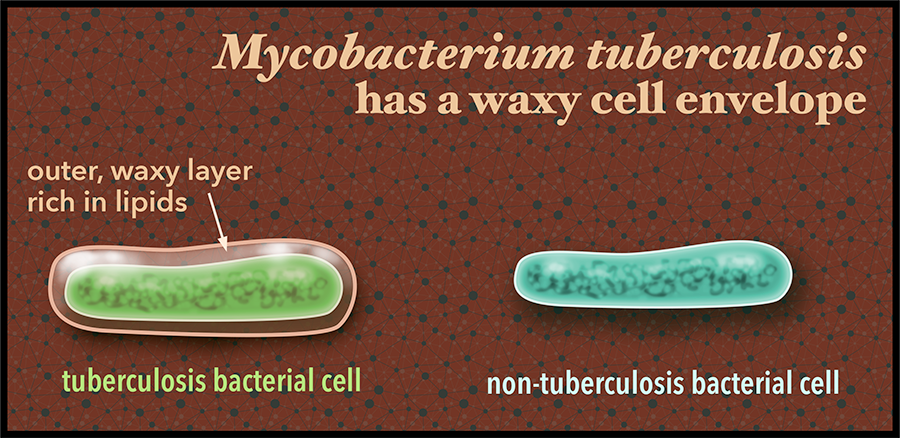
Description: The outer cell envelope of the bacteria that causes tuberculosis, Mycobacterium tuberculosis, is rich in fatty molecules called lipids. This gives the bacterial cell an unusual waxy exterior. These lipids may also play a role in conferring drug resistance to the bacteria. Tuberculosis sickened more than 9 million people across the world in 2014 and half a million people were infected with bacteria resistant to common drugs to combat the illness. Graphic: Sue Medaris
Since the researchers were interested in the evolution of the bacteria within each patient, they compared the genetic codes of the individual bacterial cells collected from each. They looked at how many letters within the code differed from one cell to the next within each patient and they looked at how much overall variation there was between each bacterial cell. From this, they derived a numerical way to describe the diversity of the bacterial populations found in each patient over time.
Most studies describe tuberculosis diversity by comparing one patient’s tuberculosis to another, so that 50 patients might represent 50 different types of tuberculosis bacteria. Pepperell calls this the “sky-level view.”
However, Pepperell says in their study they “zoomed in” and looked at the patterns of diversity of bacteria harbored by a single person with tuberculosis. Among individual patients with severe disease, they found vast diversity, even more so than between patients studied at sky-level. The findings mean there is more tuberculosis diversity within a single person with severe illness than there is tuberculosis diversity between people with the disease.
According to the World Health Organization, in 2014, 9.6 million people were sickened with tuberculosis, including 1 million children, and nearly 500,000 people developed multidrug-resistant disease. One third of the global population is infected with the tuberculosis bacteria, though only one in 10 of them will become sick with the disease in their lifetime.
The researchers also found that tuberculosis bacteria evolve substantially within their human hosts due to the pressures they face there, and that much of the genetic changes occur among the genes involved in making, regulating and transporting the cellular chemicals that form the fatty, waxy outer covering of the bacteria.
Other scientists studying everything from humans to mice have figured out that these genes interact with and manipulate the human immune system, says Pepperell. The researchers speculate the variability and redundancy in these cell envelope genes provide flexibility for the bacteria to rapidly adapt and become drug resistant.
This may present potential targets for new drugs in the future, or even vaccines to prevent tuberculosis infection.
With the data, Pepperell says they were also curious whether drug treatment reduced the diversity of the tuberculosis bacteria population in individual patients.
“We would hope, intuitively, that if you have a big, diverse population of bacteria and then you give treatment, the population of bacteria shrinks and there should be less diversity,” Pepperell says. “We did not see that.”
However, the team learned that as the severity of a person’s illness progressed, the diversity of their bacteria increased. “What leads to someone dying of tuberculosis is the bacterial population getting completely out of control,” Pepperell says.
New technology enabled the team to conduct the study this way, Pepperell says, and it’s allowing other researchers to survey entire bacterial populations within the expansive environment that is the individual human body.
“When you think about it, it makes sense to do this, because the human body is where a microbe does its business,” she says. It’s the environment that impacts each person’s bacterial residents most.
“If you only ever looked at one cell from this person and one cell from another person, you would be missing out on a lot of information about the environment the bacteria lives in, what it’s doing, what it’s responding to, what it has to cope with,” Pepperell says. “I try to think like a mycobacterium. What would my life be like?”




