UW-Madison researcher wins Klaus Biemann Medal
Josh Coon’s work has weight. It’s right there in the name: mass spectrometry.
It’s there in the results, too, enabling science by giving researchers an ever-sharper understanding of the molecules that make up living things.
“It greatly broadens the scope of biological questions you can ask,” Coon says.
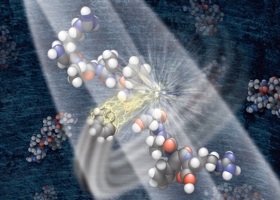
This graphic depicts a negatively charged ion breaking a protein into component parts that can be analyzed by a mass spectrometer to determine the proteins mass, structure and quantity in a sample. It’s a method Coon co-invented.
And it all comes down to mass.
“The field is about making instruments designed to measure the mass of molecules,” says Coon, professor of chemistry and biomolecular chemistry at the University of Wisconsin–Madison. “From that, you can figure out all kinds of things about them: their formation, the structure, the amount you have. You can get really creative once you have mass, and that all started with Klaus Biemann in the 1960s.”
Coon served as a post-doctoral researcher in the University of Virginia lab of Donald Hunt, co-inventing electron transfer dissociation, a method of mass spectrometry. Hunt did his post-doctoral work in Biemann’s lab at Massachusetts Institute of Technology.
The American Society for Mass Spectrometry has tightened the academic family circle by awarding Coon its Klaus Biemann Medal for “significant achievement in basic or applied mass spectrometry made by an individual early in his or her career.”
In a sense, mass spectrometers throw molecules at a wall — slicing them up with streams of electrons and bending their path with radiofrequency fields — and record the masses of the pieces. Just how far off their original path the component parts are bumped and pulled reveals how much the molecule weighed.
Coon is best known for his contribution to electron transfer dissociation, which substitutes the electrons with a stream of anions, small molecules carrying extra electrons.
“ETD allows you to characterize different types of proteins, and works well for types of proteins that mass spec isn’t typically good with,” says Coon, who continues to improve ETD in his UW–Madison lab. “We grind up a cell and look at what’s in it. It’s shotgun proteomics.”
About 500 mass spectrometry machines around the world incorporate ETD to extend their capabilities. In 2011, there were 140 published scientific studies that made direct use of the technology.
That may be the long way to measure the impact of Coon’s research. Asking collaborator Alan Attie is the short route.
“Without him, without what his work allows us to do?” asked Attie, a UW–Madison biochemist studying the connections between obesity and Type 2 diabetes, “I think I would be crying.”
Attie calls Coon as a pioneer in “phosphoproteomics,” the study of the way addition and subtraction of phosphorus atoms help or hinder complex proteins that drive so much of the necessary work inside living cells.
“A lot of the signaling pathways in us do what they do because they stimulate a cascade of events in cells that attach or detach phosphorus from proteins,” Attie says. “What Josh has done is develop techniques that survey the thousands of proteins in a cell to find which ones have phosphorus and which don’t, and go even deeper to find which particular amino acid piece of the protein is the attachment point for phosphorus.”
Attie’s lab work has progressed down lines of inquiry they would not even have considered without Coon’s enlightening mass spectrometry methods.
“He was essential in finding a signaling pathway in mice that determines whether an obese mouse gets Type 2 diabetes or not,” Attie says. “We can’t even look there without Josh, much less find anything.”
Enabling more research like Attie’s is the goal for Coon.
“Now we are working to on combining ETD with other new technologies and ideas to extend the medical impact of mass spectrometry by monitoring protein differences across hundreds or even thousands of people.”
Simply lining up 100 mass spectrometers won’t do, he says.
“There’s a lot of variability in the things we see in each experiment,” Coon says. “You need both the throughput to handle tissue samples from an enormous number of people and the repeatability to make sure you can compare all those results.”
It’s the steps that led from Biemann’s lab into Hunt’s lab and Coon’s lab — and into labs around the world studying how to tackle outstanding biomedical problems — that will be foremost in Coon’s mind when he accepts the Biemann Medal at the ASMS annual conference later this month.
“I want to talk a bit about my perspective on the people who do this work,” Coon says. “There are a lot of people who make your group what it is, and they should all share in the success.”
Tags: biosciences, research



